Data from: Allostery through the Computational Microscope: cAMP Activation of a Canonical Signalling Domain
Data from: Allostery through the Computational Microscope: cAMP Activation of a Canonical Signalling Domain
About this collection
- Extent
-
1 digital object.
- Cite This Work
-
Malmstrom, Robert D.; Kornev, Alexandr P.; Taylor, Susan S.; Amaro, Rommie E. (2015). Data from: Allostery through the computational microscope: cAMP activation of a canonical signalling domain. UC San Diego Library Digital Collections. https://doi.org/10.6075/J0Z60KZS
- Description
-
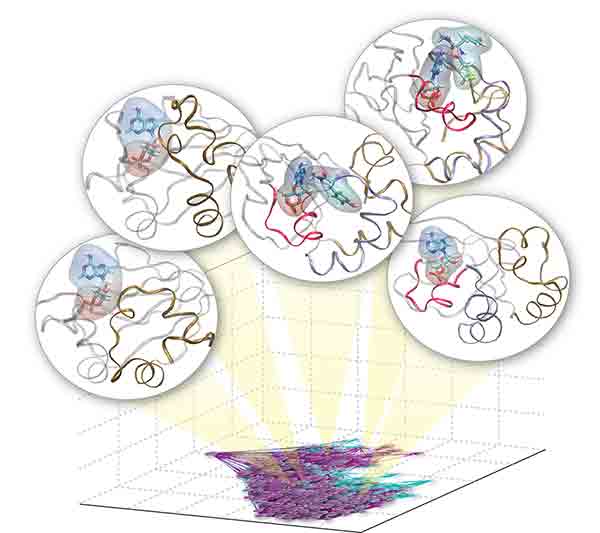
This collection contains the molecular dynamics trajectories scripts and models used in the paper “Allostery through the computational microscope: cAMP activation of a canonical signalling domain”. The purpose of this collection is to share the simulation data used to build the models and to show how the models presented in the paper were constructed and analyzed.
- Creation Date
- 2014-08-24
- Date Issued
- 2015
- Authors
- Co Principal Investigators
- Note
-
Funding information: This work was funded in part by the National Institutes of Health (NIH) through the NIH Director’s New Innovator Award Program (DP2-OD007237 to REA, R01-GM034921 to S.S.T.) and an NSF XSEDE supercomputer resources grant (RAC CHE060073N to R.E.A.). Funding from the National Biomedical Computation Resource (NBCR), NIH P41 GM103426 from the National Institutes of Health, is gratefully acknowledged. Anton computer time was provided by the Pittsburgh Supercomputing Center (PSC) and the National Center for Multiscale Modeling of Biological Systems (MM Bios) (grant P41GM103712-S1 from the National Institutes of Health). D.E. Shaw Research generously made the Anton machine at PSC available.
- Topics
Formats
View formats within this collection
- Related Resources
- Malmstrom, R. D., Kornev, A. P., Taylor, S. S., Amaro, R. E. (2015). Allostery through the computational microscope: cAMP activation of a canonical signalling domain. Nature Communications 6:7588. https://doi.org/10.1038/ncomms8588
- Assisted Model Building with Energy Refinement (Amber) (ambermd.org)
- MSMBuilder 2.51 (msmbuilder.org)
- Visual Molecular Dynamics (VMD): https://www.ks.uiuc.edu/Research/vmd/
Primary associated publication
Software
 Library Digital Collections
Library Digital Collections