UC San Diego faculty, staff and students now have full access to BioCyc, a collection of more than 14,500 Pathway/Genome databases, providing a reference on genomes and metabolic pathways of thousands of sequenced organisms.
You can search by genes, proteins, enzymes, metabolic pathways, or use the structured query for a more advanced search. There are 3 tiers of databases. The Tier 1 databases, including EcoCyc, MetaCyc, and HumanCyc are manually managed and frequently updated. The initial search defaults to Escherichia coli K-12 substr. MG1655 (EcoCyc), but you can easily switch to one of the other 14,500+ databases.
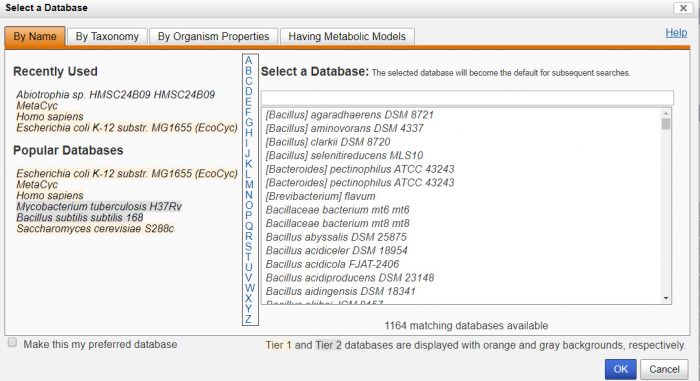
Support resources include a guided tour, website user guide, webinars, and a guide to the BioCyc database collection.
BioCyc PGDBs are generated by software that predict the metabolic pathways of completely sequenced organisms, predict which genes code for missing enzymes in metabolic pathways, and predict operons. BioCyc also integrates information from other bioinformatics databases, such as protein feature and Gene Ontology information from UniProt. The BioCyc website provides a suite of software tools for database searching and visualization, for omics data analysis, and for comparative genomics and comparative pathway questions.

