Data from: Multiscale Simulations Examining Glycan Shield Effects on Drug Binding to Influenza Neuraminidase
Data from: Multiscale Simulations Examining Glycan Shield Effects on Drug Binding to Influenza Neuraminidase
About this collection
- Extent
-
1 digital object.
- Cite This Work
-
Seitz, Christian; Casalino, Lorenzo; Konecny, Robert; Huber, Gary; Amaro, Rommie E.; McCammon, J. Andrew (2020). Data from: Multiscale Simulations Examining Glycan Shield Effects on Drug Binding to Influenza Neuraminidase. UC San Diego Library Digital Collections. https://doi.org/10.6075/J04J0CN8
- Description
-
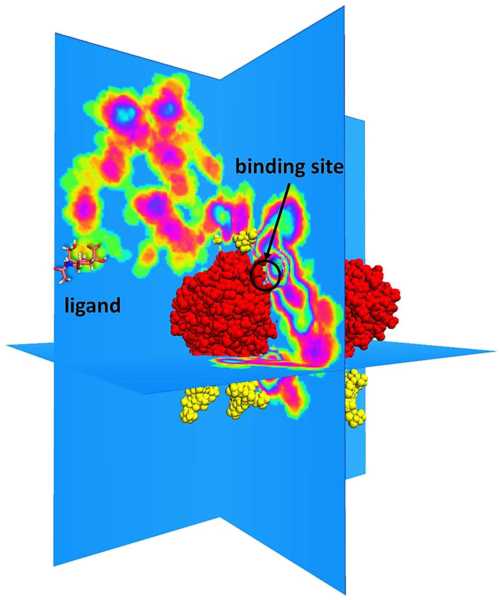
Abstract: Influenza neuraminidase is an important drug target. Glycans are present on neuraminidase and are generally considered to inhibit antibody binding via their glycan shield. In this work, we studied the effect of glycans on the binding kinetics of antiviral drugs to the influenza neuraminidase. We created all-atom in silico systems of influenza neuraminidase with experimentally derived glycoprofiles consisting of four systems with different glycan conformations and one system without glycans. Using Brownian dynamics simulations, we observe a two- to eightfold decrease in the rate of ligand binding to the primary binding site of neuraminidase due to the presence of glycans. These glycans are capable of covering much of the surface area of neuraminidase, and the ligand binding inhibition is derived from glycans sterically occluding the primary binding site on a neighboring monomer. Our work also indicates that drugs preferentially bind to the primary binding site (i.e., the active site) over the secondary binding site, and we propose a binding mechanism illustrating this. These results help illuminate the complex interplay between glycans and ligand binding on the influenza membrane protein neuraminidase.
- Creation Date
- 2017 to 2020
- Date Issued
- 2020
- Principal Investigators
- Research Team Head
- Analysts
- Funding
-
This work was supported in part by grants from the National Institutes of Health, USA (NIH grant T32EB009380 to C.S. and GM031749 to J.A.M. and G.H.). This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650112 to C.S. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) Comet at the San Diego Supercomputer Center (SDSC) through allocation csd373 to R.E.A.
- Topics
Format
View formats within this collection
- Language
- No linguistic content; Not applicable
- Identifier
-
Identifier: Christian Seitz: https://orcid.org/0000-0002-5159-8896
Identifier: Gary Huber: https://orcid.org/0000-0002-5936-6184
Identifier: J. Andrew McCammon: https://orcid.org/0000-0003-3065-1456
Identifier: Lorenzo Casalino: https://orcid.org/0000-0003-3581-1148
Identifier: Robert Konecny: https://orcid.org/0000-0002-8017-2915
Identifier: Rommie E. Amaro: https://orcid.org/0000-0002-9275-9553
- Related Resources
- Seitz, C., L. Casalino, R. Konecny, G. Huber, R. Amaro, J. A. McCammon, 2020. Multiscale Simulations Examining Glycan Shield Effects on Drug Binding to Influenza Neuraminidase. Biophys. J. https://doi.org/10.1016/j.bpj.2020.10.024
- Press coverage - ”Clearing the Course for Glycans in Development of Flu Drugs”: https://ucsdnews.ucsd.edu/feature/clearing-the-course-for-glycans-in-development-of-flu-drugs
Primary associated publication
Other resource
 Library Digital Collections
Library Digital Collections