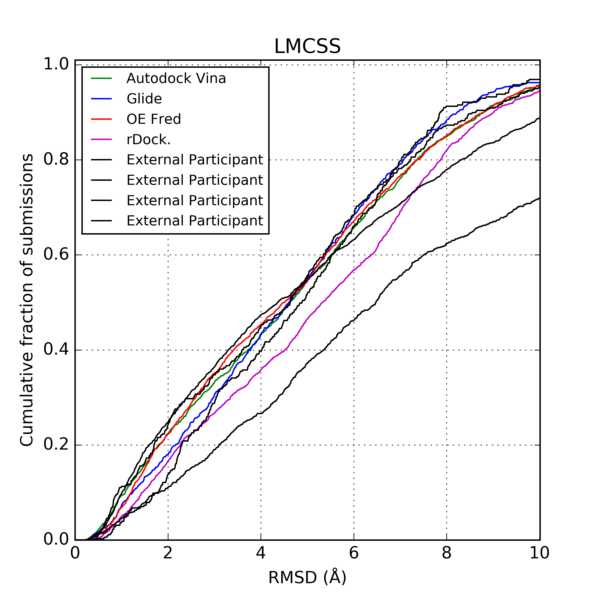
Data from: Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking
Data from: Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking
About this collection
- Extent
-
1 digital object.
- Cite This Work
-
Wagner, Jeffrey R.; Churas, Christopher P.; Liu, Shuai; Swift, Robert V.; Chiu, Michael; Shao, Chenghua; Crawl, Daniel; Feher, Victoria A.; Burley, Stephen K.; Gilson, Michael K.; Amaro, Rommie E. (2019). Data from: Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking. UC San Diego Library Digital Collections. https://doi.org/10.6075/J0610XPS
- Description
-
All structures and RMSDs used in Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking are provided here. Note that:
- Predicted structures and calculated RMSD values are available from 2017 week 5 to 2018 week 28, however several weeks in this interval were excluded from the analysis, as less than three predictions were submitted for any target.
- 2017 week 11 is excluded from the analysis as it had ~300 nearly-identical complexes released.
- The files contained in this repository are grouped by submitter, but have had comment fields scrubbed of information to preserve the anonymity of external participants.
- The settings and software environment of the GLIDE participant were changed during the "live" data collection period, so all GLIDE predictions were regenerated for this paper using Schrodinger suite version 2015-3 and the code at https://github.com/drugdata/d3r_in_house_glide as of commit fb38a3a.
- Sporadic automation and infrastructure issues prevented predictions from being generated or evaluation from being run for some targets. Notably, GLIDE predictions in 2018 between weeks 1-9 had a configuration error and did not generate RMSDs.
- External participants enrolled in CELPP at different times, were not required to submit predictions each week, and were not required to use the same method throughout the data collection period. - Date Collected
- 2017 to 2018
- Date Issued
- 2019
- Authors
- Principal Investigators
- Contributor
- Technical Details
-
The zipped workflows all require installation of the "d3r" python package via pip. Several further require installation of UCSF Chimera, babel, MGLTools, Schrodinger Suite, or the OpenEye Toolkits. Test files and a test run script are provided within each workflow's test_data directory.
Software versions: Chimera 1.10.1, RDKit 2016.3.3, MGLTools 1.5.7, AutoDock Vina 1.1.2, Schrodinger 2015-3 release, Omega 2.5.1.4, FRED 3.0.1, RBDock 2013.1/901 - Funding
-
D3R is supported by NIH grant U01 GM111528 to REA and MKG.
- Topics
Formats
View formats within this collection
- Language
- English
- Related Resources
- Jeffrey R. Wagner, Christopher P. Churas, Shuai Liu, Robert V. Swift, Michael Chiu, Chenghua Shao, Victoria A. Feher, Stephen K. Burley, Michael K. Gilson, Rommie E. Amaro (2018) Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking. bioRxiv. https://doi.org/10.1101/469940
- Jeffrey R. Wagner, Christopher P. Churas, Shuai Liu, Robert V. Swift, Michael Chiu, Chenghua Shao, Victoria A. Feher, Stephen K. Burley, Michael K. Gilson, Rommie E. Amaro (2019) Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking. Structure. https://doi.org/10.1016/j.str.2019.05.012
- Scoring code: https://zenodo.org/record/2578191#.XLkHQqZlDOQ
- Drug Design Data Resource website: https://drugdesigndata.org/
Primary associated publication
Software
Other resource
 Library Digital Collections
Library Digital Collections